Coronavirus MPro protease
The content of this page was edited by matteoferla on the 2021-10-20 20:13:18.944289.The administrators of this site take no legal responsibility for the content of this page, if you believe this page is in violation of the law, please report it.
Description
Since the release of the genome of the new coronavirus (2019-nCoV), there has been a flurry of open activity both on the vaccine front and on the drug design front, including several clinical trials to repurpose several existing drugs. In terms of structural biology, the structure of the 3C-like protease was released on the 5th February and was quickly followed by several other structures.
Protease
The most studied of the coronavirus enzymes is most definitely the protease 3CL (also known as MPro). Here shown with its substrate (protease from PDB:6Y2E and peptide TSAVLQSGFRK from SARS protease PDB:2Q6G).
Role
To save genetic space, many RNA viruses employ a strategy wherein different protein acting at the same stage of the infection cycle are produced as a single long peptide that gets cut up by a protease at given recognition sequences. In the case modelled here, between a glutamine and a serine.
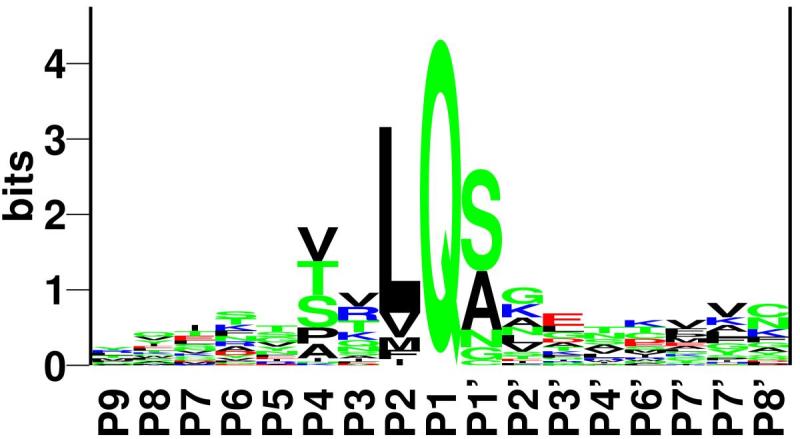 (figure from Kiemer et al. 2004)
(figure from Kiemer et al. 2004)
Activity
A protease works thanks to a catalytic triad: an acid hydrogen bonds to a base, which in turns deprotonates a nucleophile making it more active. In the case of viral protease the acid is part of the ligand, so it's a dyad. With 2019-nCoV (and SARS) histidine-163 is the base and cysteine-145 is the nucleophile.
The cysteine attacks the backbone ketone of the glutamine of the substrate. The tetrahedral hemiacetamide intermediate is highly unstable and the N-terminal half of the peptide leaves. The thioester between the enzyme and the intermediate is more stable but undergoes a substition reaction with water (hydrolysis) freeing the enzyme for another round of catalysis and releasing the second half of the substrate.
Inhibitors
Given the following:
- it is a very essential protein, so inhibiting it will stop the infection
- its substrate is large, which means it has an exposed active site to accommodate its substrate, so drug design is easier
- its substrate is a long peptide, so it is easy to make peptidomimetic drugs.
It is clear why viral protease made good candidates. In fact, the crystallisation of the HIV protease was a big deal —appearing the Voet and Voet textbook and featuring in the book The Billion-Dollar Molecule and there are many HIV protease inhibitors on the market (NB. HIV protease has a diffent structure but works on the same principle).
N3
The first structure (PDB:6LU7) of the 2019-nCoV protease was released with the candidate drug called "N3" found by the same authors in a SARS protease drug screen (Yang et al., 2005) —the 2019-nCoV protease is 96% identical to SARS protease (also a coronavirus). How it works is that the reaction starts, but gets stuck because the substrate is not quite the same. First, the nucleophilic cysteine attacks the substrate, but, instead of attacking a ketone of a peptide bond like a normal hydrolysis, it goes for the more electrophilic double bond next to it, which therefore acts as a Michael acceptor, forming a very stable, so goes nowehere and the enzyme is broken!
In terms of the structure of the peptide N3, reading it from the N-terminus (biochemistry way, as opposed to chemistry way): N-terminal cap - alanine - valine - leucine - Michael acceptor amino acid analogue with a oxidanylidenepyrrolidine side chain - C-terminal cap. The AVL part is the same as the consensus sequence, but then instead of a glutamate there is an amino acid of similar length that has a closed ring at the end. However, this large peptide has micromolar affinity for the protease of SARS and is very large, making it not ideal as drug (cf. Lipinski's rule of five).
alpha-ketoamide 13b
The next inhibited structure, contains what the depositors call alpha-ketoamide 13b. This consists of: N-terminal cap - an aromatic ringed backbone amino acid - cyclopropylalanine - the same oxidanylidenepyrrolidine side chain amino acid as N3 - C-terminal cap. That is cyclopropylalanine is taking the place of leucine and the more rigid cyclic amino acid is taking the place of valine. The reason for the latter is that peptides are very flexible, which makes them less drug like.
Diamond Screen
Recently Diamond have done a fragment screen against the protease. Fragment screens are great as starting data for virtual screens as the protein side chains or mobile loops may adopt different conformations potentially resulting in stronger affinity.
Mpro-x0995, Mpro-x0830, Mpro-x1012, Mpro-x0177, Mpro-x1132, Mpro-x1249, Mpro-x1311, Mpro-x0354, Mpro-x0540, Mpro-x1418, Mpro-x1308, Mpro-x1425, Mpro-x1374, Mpro-x0195, Mpro-x0678, Mpro-x0464, Mpro-x0499, Mpro-x1235, Mpro-x0390, Mpro-x0350, Mpro-x1237, Mpro-x1093, Mpro-x1392, Mpro-x0376, Mpro-x1077, Mpro-x0991, Mpro-x0478, Mpro-x1478, Mpro-x1334, Mpro-x0874, Mpro-x0831, Mpro-x0072, Mpro-x1336, Mpro-x0336, Mpro-x0828, Mpro-x1402, Mpro-x0786, Mpro-x1348, Mpro-x1420, Mpro-x0194, Mpro-x0759, Mpro-x1375, Mpro-x1458, Mpro-x1351, Mpro-x0104, Mpro-x0691, Mpro-x0734, Mpro-x1384, Mpro-x0755, Mpro-x1002, Mpro-x1386, Mpro-x0820, Mpro-x1187, Mpro-x1382, Mpro-x0887, Mpro-x1380, Mpro-x0161, Mpro-x1119, Mpro-x0305, Mpro-x0748, Mpro-x1412, Mpro-x0387, Mpro-x0805, Mpro-x0692, Mpro-x0107, Mpro-x0434, Mpro-x0770, Mpro-x0946, Mpro-x0689, Mpro-x0769, Mpro-x1385, Mpro-x0669, Mpro-x0774, Mpro-x1358, Mpro-x0749, Mpro-x0398, Mpro-x1226, Mpro-x0752, Mpro-x1493
PDB structures
Protease
- Protease in complex with an inhibitor N3 ( PDB:6LU7 )
- Protease in apo form ( PDB:6Y2E )
- Protease in complex with alpha-ketoamide 13b ( PDB:6Y2F )
- Protease in complex with alpha-ketoamide 13b ( PDB:6Y2G )
Other enzymes
There are other enzymes in 2019-nCoV that are not crystallised, most notably the RNA polymerase (nsp12), which is potently inhibited by the drug remdesivir.
Structural protein
- HR2 Domain ( PDB:6LVN )
- Post fusion core of S2 subunit ( PDB:6LXT )
- Prefusion spike glycoprotein with a single receptor-binding domain up ( PDB:6VSB )
- Spike protein complexed with receptor ( PDB:6VSJ )
- Chimeric receptor-binding domain complexed with its receptor human ACE2 ( PDB:6VW1 )
Models
the I-Tasser authors have released the predicted protein structures of all of the proteins in the viral genome.