CAD
The content of this page was edited by admin and matteoferla on the 2022-02-01 15:54:17.049046.The administrators of this site take no legal responsibility for the content of this page, if you believe this page is in violation of the law, please report it.
CAD (carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase trifunctional enzyme)
Multiple structures The structure shown is a composition of a threaded structure and two crystal structure. How they orient relative to each other is unknown.
Structure
CAD is a fusion of four different protein activities in the pyrimidine pathway in three separate domains as illustrated in this diagram from Ruiz-Ramos et al. 2016 :
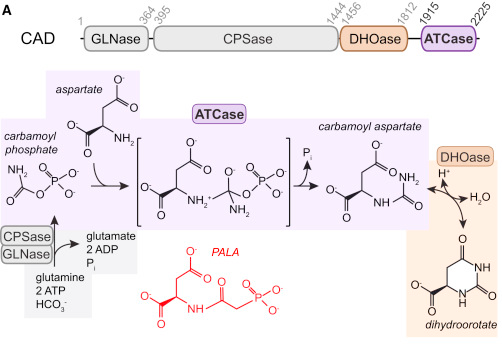
The structure here is the whole of CAD protein. It is composed of three separate structures and how they join is not known, nor is it certain how it oligomerises.
- The N-terminus is a Swiss-model based upon PDB:5DOU (56%). This acts as a glutamine-dependent carbamoyl phosphate synthetase (GLNase+CPSase)
-
The middle is PDB:4C6E, dihydroorotase (DHOase)
-
The C-terminus is PDB:5G1N as a trimer (normally a dimer of trimers), aspartate transcarbamoylase (ATCase)
Mutations
- C92R (wild type/mutant) This mutation results in a bulkier residue in the core of the protein, this will destabilise the subdomain (termed S1). It is not known what is the role of this domain (cf. crystal structure paper ). It is far (26Å) away from the ADP/ATP in the active site. In Carbamoyl phosphate synthetase I (CPS1) this residue corresponds to serine-137 (same shape). As remarked in Ruiz-Ramos et al. 2016, there are several cavities in this neighborhood: these may be allosteric binding sites, therefore, this mutation may be interfering with allosteric regulation, therefore resulting in the gain of function phenotype, however, without knowing what is the role of subdomain S1 it cannot be said.
- D2047∗ (remnant/missing). This mutation removes the aspartate transcarbamoylase functionality. The ligand present is PALA, a drug analogous to the substrates (aspartate and carbomoyl-phosphate). This chain may still oligomerise given that it has intact CPSase and DHOase domains, which would likely have either a positive or a negative on the activity of the wild type polypeptide chain in light of the cooperative activation seen in this domain (ATCase), where the domains can be in a closed state where substrate binding is harder, or in an open state where substrate binding is easier.
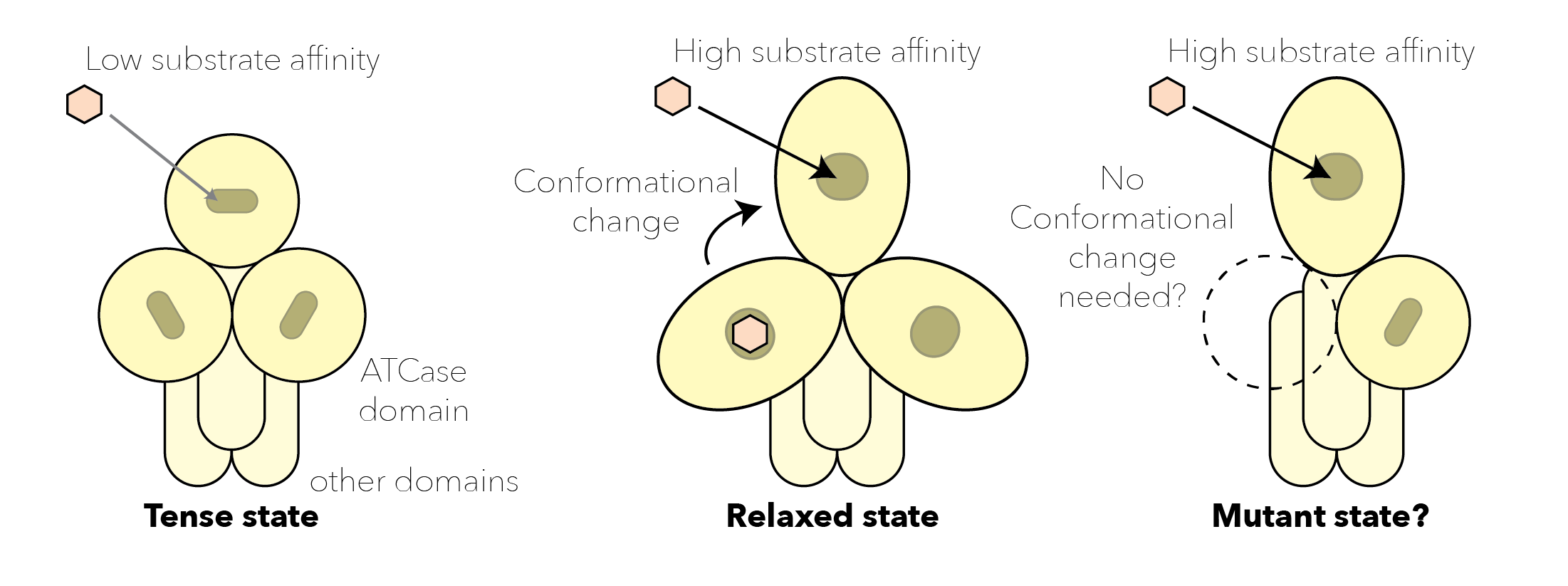
- V2115L (wild type/mutant in apo form). The mutation does not destabilise the structure in the relaxed state (induced by the cooperative binding of ligand or the drug PALA), but does in the apo-form (tense state, PDB:5G1O). This may result in a preference for the relaxed state (more active) over the tense state (less active).